Tommi Nyman Research
Molecular Ecology in Arctic and Subarctic Ecosystems

Below is a description of my ongoing and past main projects and research themes, in roughly reverse order. However, most of these broad lines of research have been (and will continue to be) explored in parallel, and have proceeded intermittently whenever time, funding, and/or or students have been available. Research outputs that are less directly connected to these main themes can be found on the Publications page.
Insects of the Forest–Tundra Ecotone (ForTunE)
The ForTunE project focuses on the diversity of insects in subarctic and arctic habitats northernmost Norway. The region represents an ecotone where Boreal taiga forest transitions into subarctic birch forest, which then gives way to tall-shrub willow tundra and open tundra on the coast. All ecosystems of the region are and will continue to be heavily impacted by climate change. To obtain a baseline for monitoring of changes in insect communities, we conducted extensive Malaise trapping campaigns in different habitats on the Varanger Peninsula through the summers of 2020 and 2021. The material has been stored frozen in alcohol, and has been partly sorted into ten taxonomic fractions, with an emphasis on so-called "dark taxa" that are known to contain a considerable amount of unknown diversity. In other subprojects, we have characterized spatial, environmental, and temporal variation in the the pollinator community of cloudberry (Rubus chamaemorus), an economically and culturally important resource in the region, and have produced a curated DNA barcode reference library for the parasitoid communities of geometrid moths that regularly undergo devastating outbreaks in the region's mountain birch forests. The ForTunE project was funded 2019–2023 by Artsdatabanken (The Norwegian Biodiversity Information Centre), and has also been supported by the Norwegian Barcode of Life (NorBOL) network.
RemoTnitor
The RemoTnitor project aims at developing efficient methods for non-invasive monitoring of brown bear, moose, and semidomestic reindeer populations based on genetic analyses of feces collected from the field. Feces and droppings of wildlife are a treasure trove of information, because they contain DNA of the animal itself, the plants and/or animals it has consumed, as well as the parasites, pathogens, and microbes residing within its digestive system. By leveraging modern high-throughput sequencing approaches, it is therefore possible to track individuals and estimate overall population size, and to assess individual and population-level diet and health status without seeing or disturbing the animals themselves. The first line of the project focuses on development of efficient GT-seq markers for individual identification in the three focal species based on SNP datasets generated using genotyping chips and 3RAD sequencing. The second line of investigation aims at refining multiplexed metabarcoding protocols for simultaneous assessment of diet, parasites and pathogens, and gut microbiomes. The third line explores whether all of the aforementioned pieces of information can be retrieved from sequence reads generated through metagenomic shotgun sequencing. RemoTnitor is a three-year collaborative project bringing together researchers from NIBIO Svanhovd, the Norwegian Veterinary Institute, UiT – The Arctic University of Norway, and the Norwegian Institute for Nature Research (NINA). The project is funded by the Norwegian Ministry of Climate and Environment and the FRAM Centre, with additional support provided by NIBIO.
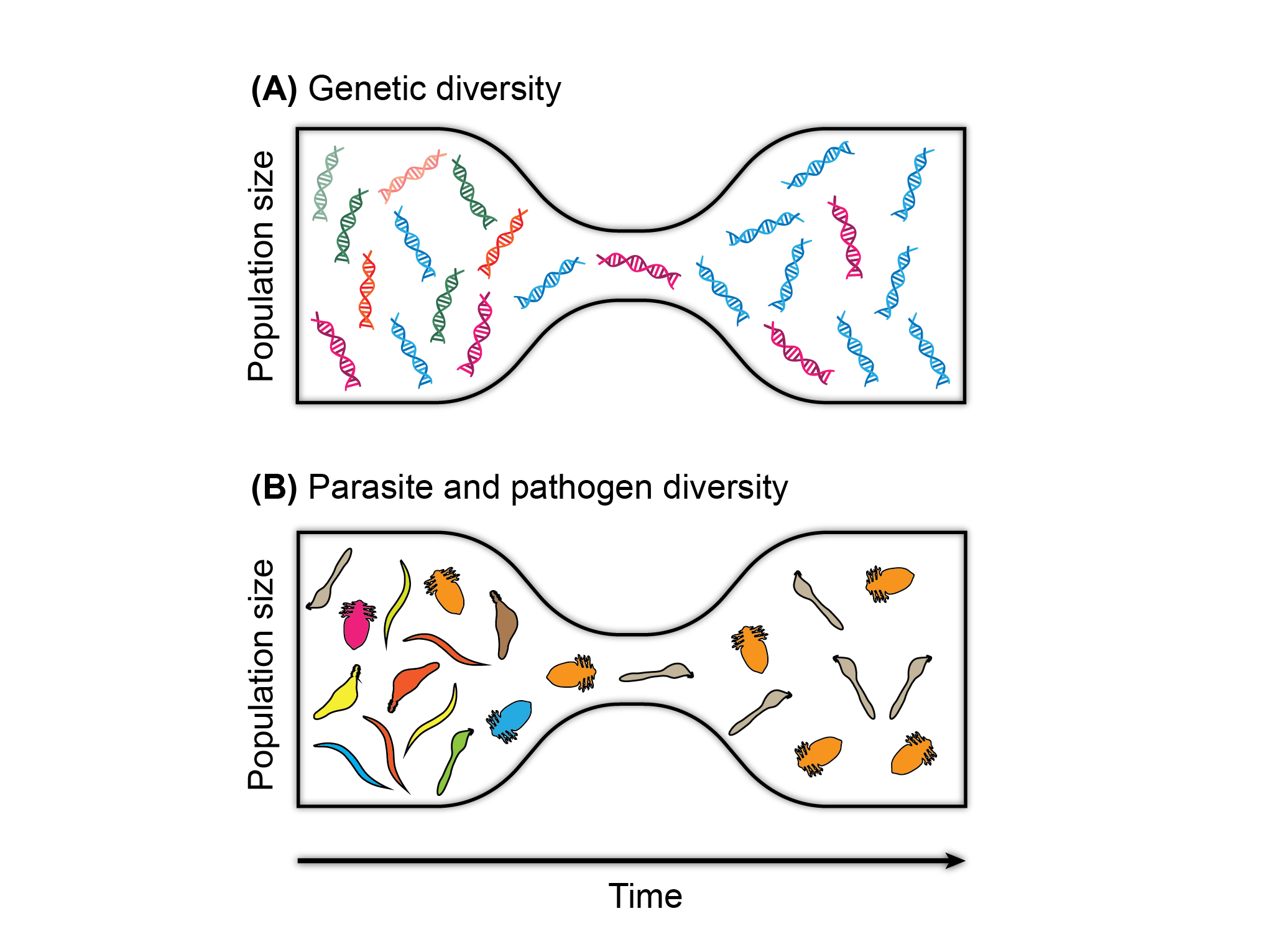
Multitrophic conservation genetics
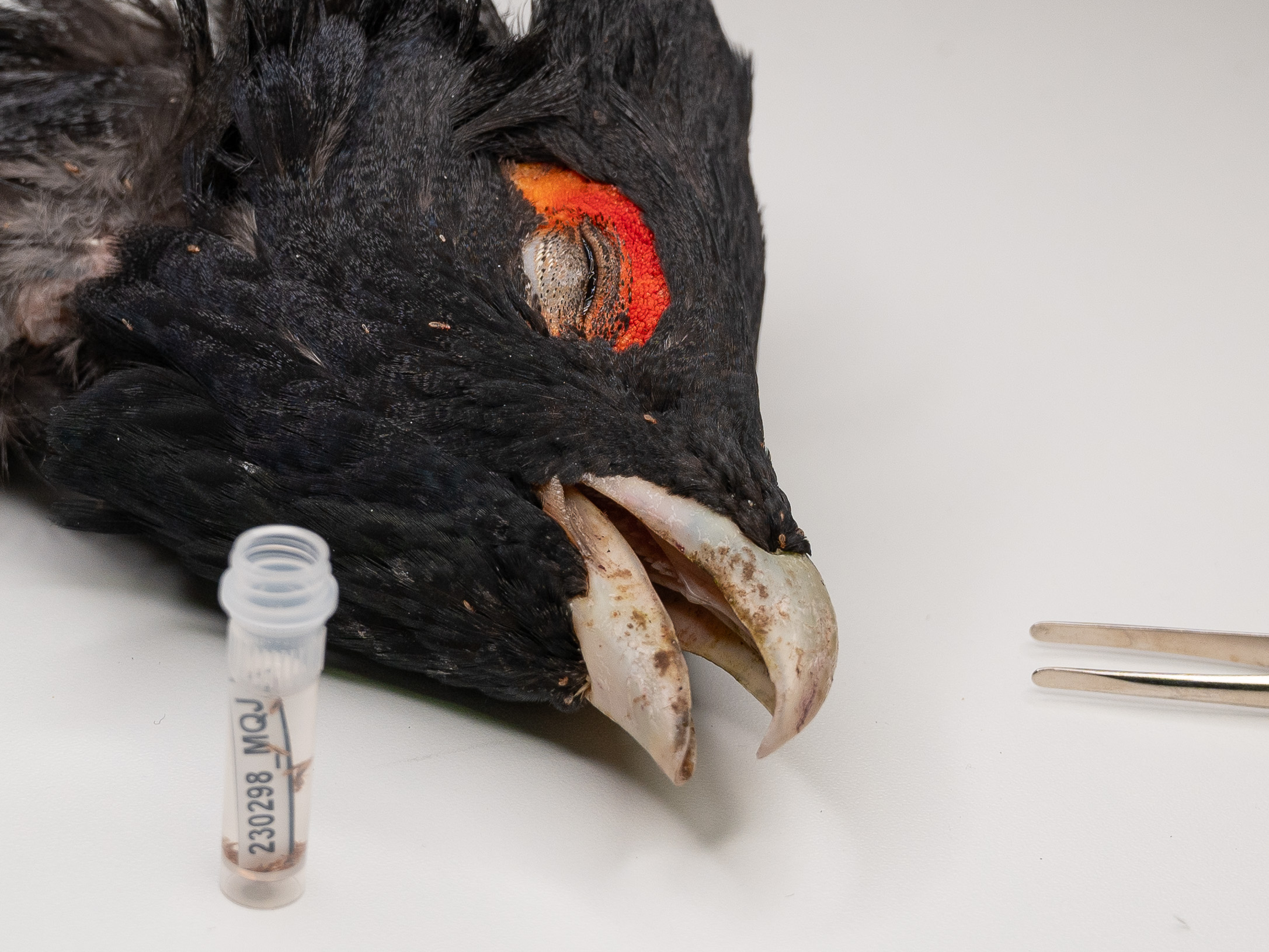
Overexploitation and habitat loss threaten numerous species across the world. Stochastic factors and inbreeding during anthropogenic population bottlenecks often reduce genetic diversity within endangered species. Such genetic erosion poses an additional threat in itself, and may impede subsequent population recovery and adaptation to changing environmental conditions. Conservation-genetic research has thus far mainly focused on large, charismatic vertebrate species. However, a parallel but largely unnoticed loss of species and genetic diversity is underway also in their specialist parasites. A multitrophic approach in conservation genetics is therefore urgently needed in order to fully appreciate the consequences of population bottlenecks. In our studies, we have focused on genomic analyses of external and internal parasites of northern European seal species and populations. Especially different ringed seal subspecies have widely divergent population sizes and levels of genetic diversity, which makes it possible to test hypotheses on how host population size and isolation influence species and genetic diversity in associated parasites. We have also used genome-level data on specialist Echinophthirius seal lice to reconstruct the postglacial history of northern European seal species and populations. The parasite studies have been done in collaboration with researchers from the University of Eastern Finland, the University of Illinois, and the University of Gdansk, and have supported by grants from many private foundations in Finland as well as the Polish Academy of Sciences (though a grant awarded to L. Sromek).
Structure and function of plant–herbivore–parasitoid food webs
The main part of biodiversity on Earth is made up of plants, plant-feeding insects, and insect parasitoids. Understanding the structure and function of multitrophic plant–herbivore–parasitoid networks constitutes a major challenge for ecological research, but is also fundamentally important for practical applications in forestry and agriculture. However, investigating associations within species-rich multitrophic food webs – and elucidating the factors that drive variation in network composition – are complicated by the sheer number and small size of species involved in these networks. Today, genetic tools such as DNA barcoding provide effective means for identifying herbivores and parasitoids. Particularly for parasitoids, an important benefit is that barcoding can be used for identifying larvae and eggs. Molecular identities can then be linked to species names whenever a reference barcode library based on morphologically identified adults is available. In my research, I have used barcode-based identification for quantifying host–parasitoid associations in willow-galling sawflies, in studies focusing on phylogenetic, spatial, and environmental drivers of parasitism. In addition, I have used phylogeneny-based tests for inferring the role of host plant and other ecological factors in host–parasitoid associations in willow-galling and leaf-mining sawflies. For fungus-associated insect communities, we have used metabarcoding of insect DNA extracted from whole fungal fruiting bodies (mushrooms) for testing the effects of host taxon and phylogeny, phenology, location, and latitude on the diversity and structure of the communities.
Drivers of diversification in plant–herbivore and host–parasite networks
Insects feeding on plants drive the main ecological processes in practically all terrestrial environments. The extreme species diversity of insect herbivores may be explained by host specialization: most insects are associated with a single host-plant species, or a group of related species. Shifts among available plant species and taxa could therefore be a major factor underlying the diversification in plant-feeding insects. In my studies, I have focused on eco-evolutionary research questions spanning both micro- and macroevolutionary time scales, and on combining genetic, ecological, geographical, and chemical data. In my studies on connections between speciation and specialization, I have mainly focused on population-genetic and population-genomic analyses of sawflies inducing galls on different willow (Salix) species. At the other extreme, I have investigated the diversificatation of ancestrally herbivorous lineages within the order Hymenoptera through the last 280 million years based on phylogenetic and phylogenomic datasets. Interestingly, plant-feeding insects are in an eco-evolutionary sense typical parasites, meaning that directly analogous questions on the role of host shits as as a driver of speciation arise when dealing with ecto- and endoparasites of mammals and birds. We have therefore used similar approaches and genomic markers for studying host-associated genetic differentiation in Corynosoma helminth worms and Echinophthirius lice associated with different seal species in northern Europe. The various plant–insect research projects have been done in collaboration with numerous researchers (see Publications), and have through the years received funding from the Academy of Finland, the Kone Foundation, and other private Finnish foundations.